Presented at the 55th ASMS Conference on Mass Spectrometry, June 4, 2007, Indianapolis, Indiana

Goal
Develop a procedure for peer review of Wikipedia articles on mass spectrometry.
Introduction
Wikipedia is an on-line encyclopedia that anyone can edit. The site is maintained by hundreds of users who create, edit and organize articles, oversee quality and consistency, moderate disputes, and guard against abuse. Anyone can contribute as much or as little as he or she desires. On the English language Wikipedia in early 2007, there were 1.4 M articles being edited by more than 150,000 users who were creating about 2000 new articles each day. Of these articles, 70 were in the category “Mass Spectrometry.”
Why is Wikipedia Important?
The public face of mass spectrometry is determined by the information that can be found most easily on the web.
Content Development Models
In April 2007, the top ten hits for “mass spectrometry” on the search engine Google were
- Wikipedia
- ASMS (What is MS?)
- ASMS
- Michigan State University
- University of Arizona
- University of Leeds
- University of Illinois, Chicago
- Scripps Institute
- Ionsource.com
- Imass.com
and on Yahoo
- Wikipedia
- I-mass.com
- Basepeak.com
- Encyclopaedia Britannica
- ASMS
- Michigan State University
- Wikipedia
- GenomicGlossaries.com
- Mass Spectrometry Blog
- University of New Mexico
Wikipedia is the #1 hit for “Mass Spectrometry” on the top five search pages
Google
Yahoo
Ask.com
MSN Live Search
Altavista
Wikipedia is the way that the public learns about mass spectrometry on the web yet the information is not subject to peer review and it is not developed in conjunction with any mass spectrometry organization.
Content Development Models
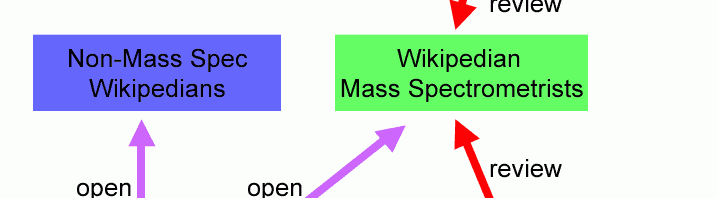
Three basic content development models are presented with their advantages and disadvantages. Here, a Wikipedian is someone who understands the workings of Wikipedia and can create and edit content and work with Editors, Bureaucrats and Administrators. A mass spectrometrist is a person who has or is working toward an advanced degree in mass spectrometry or has the equivalent work experience.
Open Model
The Open Model is in effect today. Wikipedians, who may also be mass spectrometrists, openly edit mass spectrometry content on Wikipedia, drawing on mass spectrometry literature and web-based resources.

Advantages and Disadvantages
The advantage of the open model is that it is already in place. There are mass spectrometry entries on Wikipedia and anyone can add content within the bounds of the Wikipedia community. Wikipedians with a knowledge of mass spectrometry, chemistry, physics and other areas provide an ad hoc peer review.
The disadvantages are the time required to learn the Wikipedia system to effectively generate content and the fact that the site tends to favor the persistent and tactically adroit over the subject expert. Further, content generated on Wikipedia is not credited to the student or academic and copyright is not retained.
Closed Model
In the closed model, content is generated by mass spectrometrists and in many cases subject to peer review. For example, the ASMS “What is Mass Spectrometry?” web page. Another example is mass spectrometry journals, all of which are on line and some of these are openly available.
Advantages and Disadvantages
The advantage of the closed model is that the content is created by experts and peer reviewed. The disadvantage is that the information is not generally available and is often presented at a level not suited to a general audience. Some excellent works have been generated in recent years, such as the documents created by the ASMS Education Committee, Michael A Grayson’s Measuring Mass: From Positive Rays to Proteins, and a wide range of excellent mass spectrometry textbooks. However, only a few of these works are openly available. Consider a high school student writing a report on mass spectrometry. Where will that student obtain the information for that report?
The Protected Fork Model
The material in Wikipedia is not copyrighted and it can therefore be “forked” – all or part of the material can be removed and independently modified as long as the derivative material is also made available with the same unrestricted license. This is the kind of reciprocal license principle that is applied to open source software.
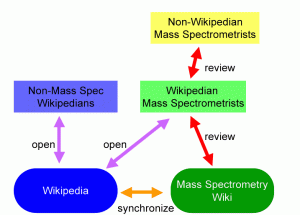
A Wikipedia fork must be available by unrestricted license, but it need not be openly edited: the fork can be protected. Under this model, mass spectrometry content is taken from Wikipedia under the terms of the GNU Free Documentation License. This “Mass Spectrometry Wiki” can only be modified by a set of approved mass spectrometry editors whose work is reviewed by their peers. Non-wikipedian mass spectrometrists can participate without the need to learn the rules and language of generating Wikipedia content. Wikipedian mass spectrometrists can help to synchronize (or at least harmonize) the content between the two wikis.
Advantages and Disadvantages
The advantage of the Protected Fork Model is that it has the potential to bring in experts on a subject who may not otherwise be inclined to participate directly in Wikipedia. It offers the possibility of peer review within the Wikipedia construct. The disadvantage is the difficulty in synchronizing information (in both directions) and the need for experienced Wikipedians who are also experienced mass spectrometrists.
Conclusions
Wikipedia’s presentation of mass spectrometry has become a significant part of the way the public perceives the field. The mass spectrometry community should be aware of this fact and avenues for participation in Wikipedia should be encouraged and facilitated.
Links
This material is based upon work supported by the National Science Foundation under Grant No. CHE-0415360