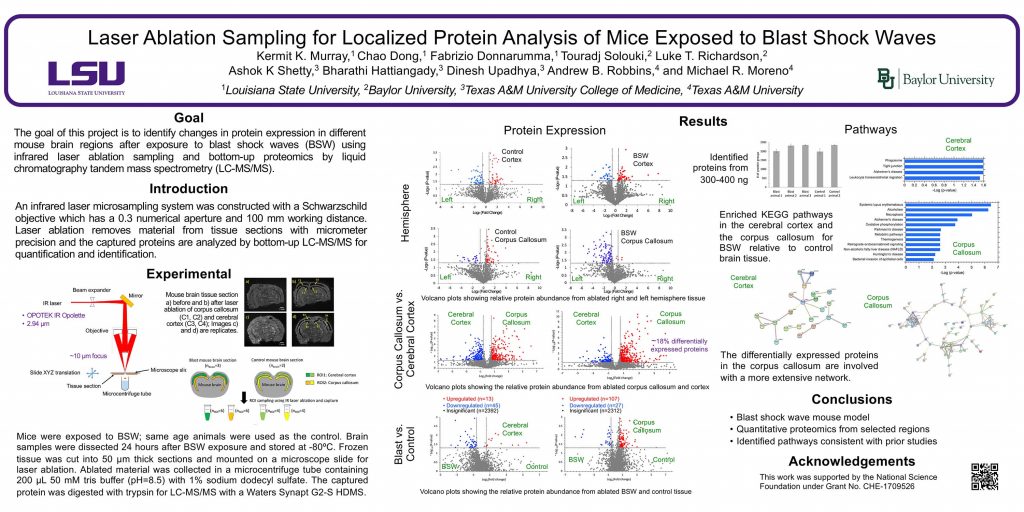
ThP 080
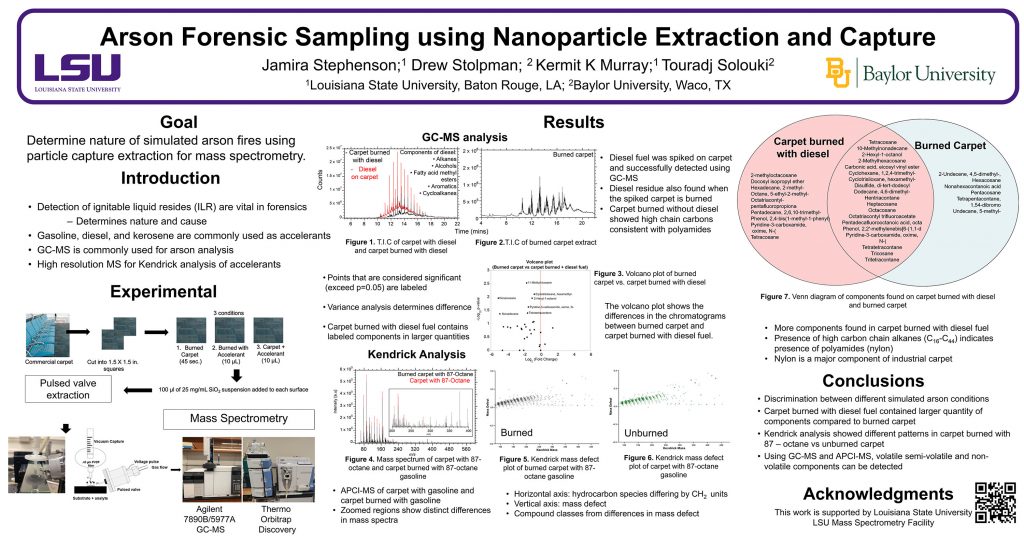
Murray Mass Spectrometry Group
Research group of Kermit Murray at Louisiana State University where we use lasers for sampling and imaging and study the chemistry and physics of laser ablation.

R.O. Lawal, L.T. Richardson, C. Dong, F. Donnarumma, T. Solouki, K.K. Murray, Deep-ultraviolet laser ablation sampling for proteomic analysis of tissue, Anal. Chim. Acta, 1184 (2021). doi: 10.1016/j.aca.2021.339021
Deep-ultraviolet laser ablation with a pulsed 193 nm ArF excimer laser was used to remove localized regions from tissue sections from which proteins were extracted for spatially resolved proteomic analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS). The ability to capture intact proteins by ablation at 193 nm wavelength was verified by matrix-assisted laser desorption ionization (MALDI) of the protein standard bovine serum albumin (BSA), which showed that BSA was ablated and captured without fragmentation. A Bradford assay of the ablated and captured proteins indicated 90% efficiency for transfer of the intact protein at a laser fluence of 3 kJ/m2. Rat brain tissue sections mounted on quartz microscope slides and ablated in transmission mode yielded 2 μg protein per mm2 as quantified by the Bradford assay. Tissue areas ranging from 0.06 mm2 to 1 mm2 were ablated and the ejected material was collected for proteomic analysis. Extracted proteins were digested and the resulting peptides were analyzed by LC-MS/MS. The proteins extracted from the ablated areas were identified and the average number of identified proteins ranged from 85 in the 0.06 mm2 area to 2400 in the 1 mm2 area of a 50 μm thick tissue. In comparison to infrared laser ablation of equivalent sampled areas, both the protein mass and number of proteins identified using DUV laser ablation sampling were approximately four times larger.
N. Mehta, S. Shaik, A. Prasad, A. Chaichi, S.P. Sahu, Q. Liu, S.M.A. Hasan, E. Sheikh, F. Donnarumma, K.K. Murray, X. Fu, R. Devireddy, M.R. Gartia, Multimodal Label‐Free Monitoring of Adipogenic Stem Cell Differentiation Using Endogenous Optical Biomarkers, Adv. Funct. Mater., (2021) 2103955; doi: 10.1002/adfm.202103955
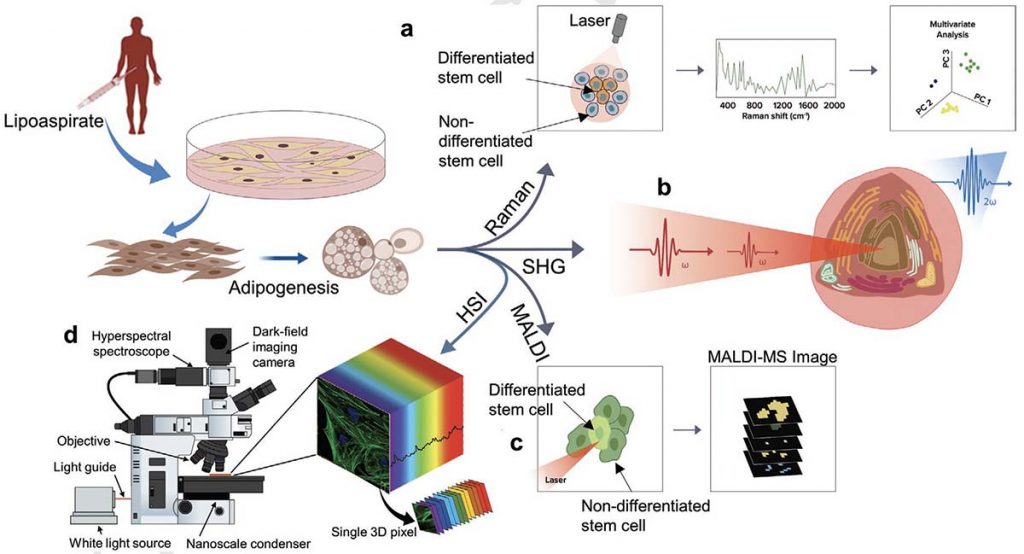
Stem cell-based therapies carry significant promise for treating human diseases. However, clinical translation of stem cell transplants for effective treatment requires precise non-destructive evaluation of the purity of stem cells with high sensitivity (<0.001% of the number of cells). Here, a novel methodology using hyperspectral imaging (HSI) combined with spectral angle mapping-based machine learning analysis is reported to distinguish differentiating human adipose-derived stem cells (hASCs) from control stem cells. The spectral signature of adipogenesis generated by the HSI method enables identifying differentiated cells at single-cell resolution. The label-free HSI method is compared with the standard techniques such as Oil Red O staining, fluorescence microscopy, and qPCR that are routinely used to evaluate adipogenic differentiation of hASCs. HSI is successfully used to assess the abundance of adipocytes derived from transplanted cells in a transgenic mice model. Further, Raman microscopy and multiphoton-based metabolic imaging is performed to provide complementary information for the functional imaging of the hASCs. Finally, the HSI method is validated using matrix-assisted laser desorption/ionization-mass spectrometry imaging of the stem cells. The study presented here demonstrates that multimodal imaging methods enable label-free identification of stem cell differentiation with high spatial and chemical resolution.
A.C. Pulukkody, Y.P. Yung, F. Donnarumma, K.K. Murray, R.P. Carlson, L. Hanley, Spatially resolved analysis of Pseudomonas aeruginosa biofilm proteomes measured by laser ablation sample transfer, PLoS One, 16 (2021) e0250911, doi: 10.1371/journal.pone.0250911
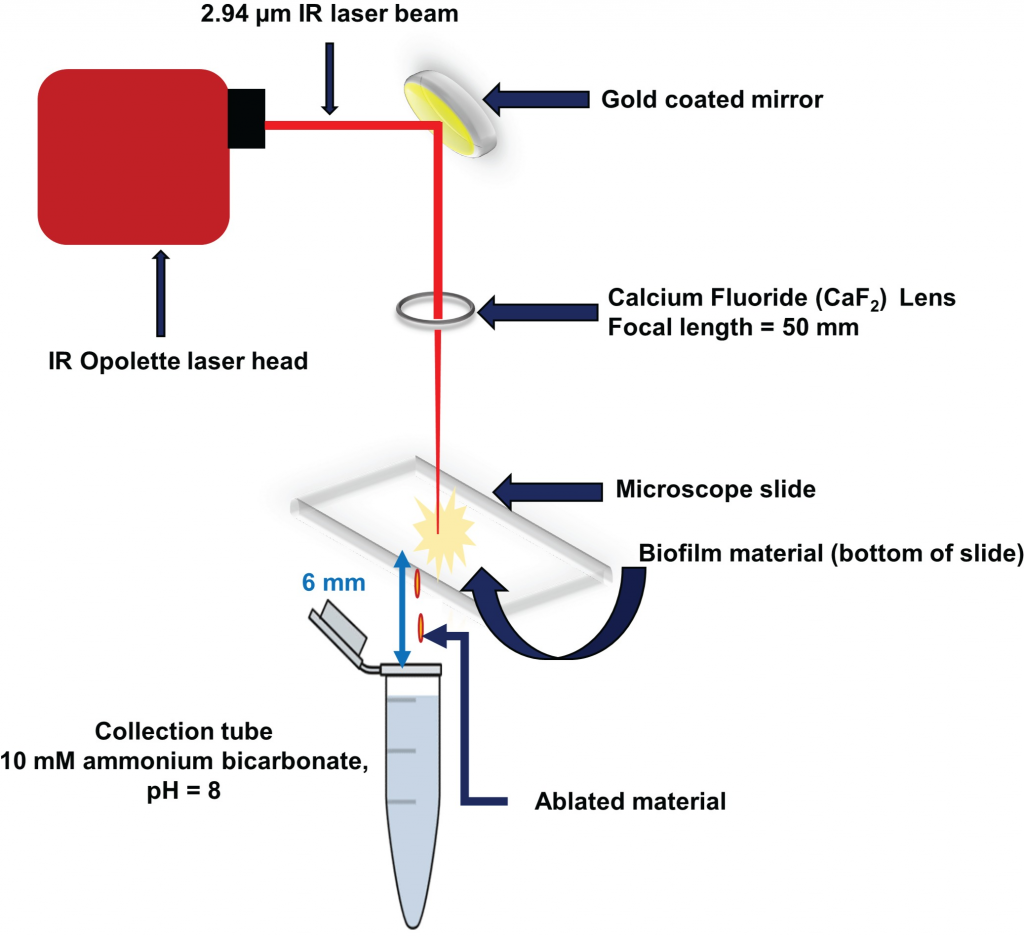
Heterogeneity in the distribution of nutrients and oxygen gradients during biofilm growth
gives rise to changes in phenotype. There has been long term interest in identifying spatial
differences during biofilm development including clues that identify chemical heterogeneity.
Laser ablation sample transfer (LAST) allows site-specific sampling combined with label
free proteomics to distinguish radially and axially resolved proteomes for Pseudomonas aeruginosa
biofilms. Specifically, differential protein abundances on oxic vs. anoxic regions of a
biofilm were observed by combining LAST with bottom up proteomics. This study reveals a
more active metabolism in the anoxic region of the biofilm with respect to the oxic region for
this clinical strain of P. aeruginosa, despite this organism being considered an aerobe by
nature. Protein abundance data related to cellular acclimations to chemical gradients
include identification of glucose catabolizing proteins, high abundance of proteins from arginine
and polyamine metabolism, and proteins that could also support virulence and environmental
stress mediation in the anoxic region. Finally, the LAST methodology requires only a
few mm2 of biofilm area to identify hundreds of proteins.
K.K. Murray, Lasers for matrix-assisted laser desorption ionization, J. Mass Spectrom., 56 (2021) e4664. https://doi.org/10.1002/jms.4664

Matrix-assisted laser desorption ionization (MALDI) was introduced 35 years ago and has advanced from a general method for producing intact ions from large biomolecules to wide use in applications ranging from bacteria identification to tissue imaging. MALDI was enabled by the development of high energy pulsed lasers that create ions from solid samples for analysis by mass spectrometry. The original lasers used for MALDI were ultraviolet fixed-wavelength nitrogen and Nd:YAG lasers, and a number of additional laser sources have been subsequently introduced with wavelengths ranging from the infrared to the ultraviolet and pulse widths from nanosecond to femtosecond. Wavelength tunable sources have been employed both in the IR and UV, and repetition rates have increased from tens of Hz to tens of kHz as MALDI has moved into mass spectrometry imaging. Dual-pulse configurations have been implemented with two lasers directed at the target or with a second laser creating ions in the plume of desorbed material. This review provides a brief history of the use of lasers for ionization in mass spectrometry and describes the various types of lasers and configurations used for MALDI.
K.K. Murray, In defense of the quasimolecular ion, J. Mass Spectrom., 56 (2021) e4700; doi: 10.1002/jms.4700.
The term quasimolecular ion has been used to describe ions comprising a molecule and weakly bound positive or negative ion or an ion formed by the loss of a proton from a molecule. This term was used in mass spectrometry from the late 1960s after the development of chemical ionization but has been deprecated in recent terms recommendations due to what is perceived as its overly broad use. This letter argues that the term is well defined and has a long history of use in mass spectrometry and other fields and should be considered as a recommended term.
V. A. Ricigliano, C. Dong, L.T. Richardson, F. Donnarumma, S.T. Williams, T. Solouki, K.K. Murray, Honey Bee Proteome Responses to Plant and Cyanobacteria (blue-green algae) Diets, ACS Food Science & Technology, 1 (2021) 17-26. 10.1021/acsfoodscitech.0c00001
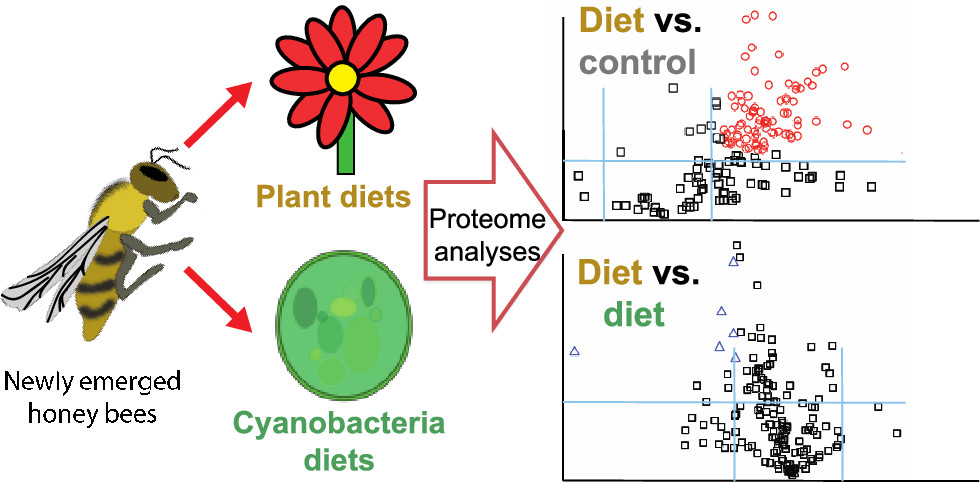
Malnutrition is an increasing threat to honey bees that can be mitigated by feeding artificial pollen substitute diets; however, little is known about the molecular mechanisms underlying their impact on bee health. Here, we examined proteomic responses to natural and artificial diets in the honey bee fat body, a tissue with central nutrient storage and metabolic functions. Bees were fed protein diets of natural pollen, a commercial plant-based diet used by beekeepers that does not contain pollen (Ultra Bee), and two novel cyanobacteria diets comprising dried or fresh laboratory-grown Arthrospira platensis (commonly, spirulina). Relative to a protein-free control group, diet consumption elicited broad upregulation of metabolic processes associated with amino acids, carbohydrates, and lipids. Plant and cyanobacteria diets led to equivalent dietary protein assimilation and a marked overlap in proteome expression patterns, indicative of comparable nutritive and metabolic impacts. This was corroborated by equivalent titers of the storage lipoprotein vitellogenin and nutritionally-regulated stress response proteins (superoxide dismutase, glutathione S-transferase 1, catalase, and heat shock protein 90). The tested diets recapitulated the proteomic effects of a natural pollen diet and support stress resistance via improved nutritional status. Our results provide new insights into the impact of artificial feed on honey bees and highlight the potential of cyanobacterial biomass as a sustainable nutrition source for improving bee health.
A. Chaichi, S.M.A. Hasan, N. Mehta, F. Donnarumma, P. Ebenezer, K.K. Murray, J. Francis, M.R. Gartia, Label-free lipidome study of paraventricular thalamic nucleus (PVT) of rat brain with post-traumatic stress injury by Raman imaging, Analyst, 146 (2021) 170-183.
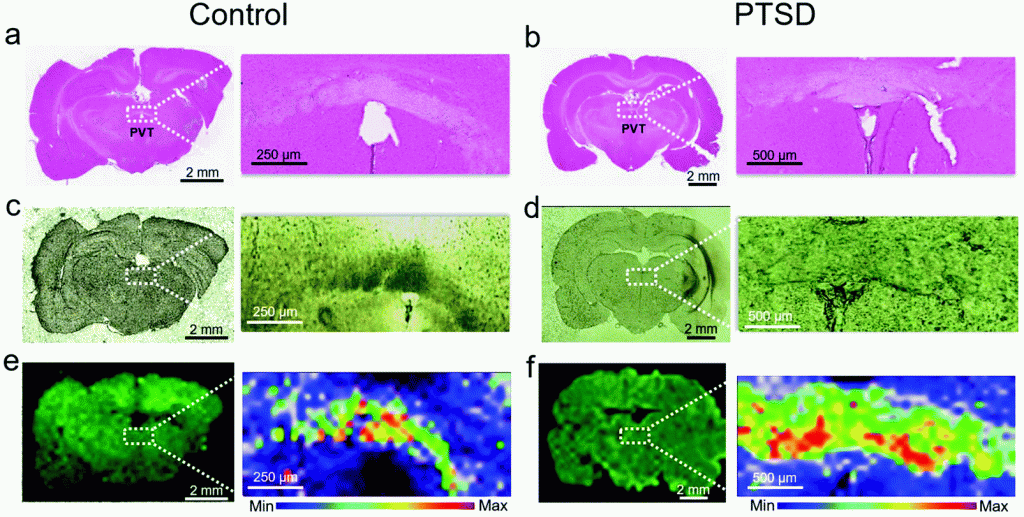
Post-traumatic stress disorder (PTSD) is a widespread psychiatric injury that develops serious life-threatening symptoms like substance abuse, severe depression, cognitive impairments, and persistent anxiety. However, the mechanisms of post-traumatic stress injury in brain are poorly understood due to the lack of practical methods to reveal biochemical alterations in various brain regions affected by this type of injury. Here, we introduce a novel method that provides quantitative results from Raman maps in the paraventricular nucleus of the thalamus (PVT) region. By means of this approach, we have shown a lipidome comparison in PVT regions of control and PTSD rat brains. Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry was also employed for validation of the Raman results. Lipid alterations can reveal invaluable information regarding the PTSD mechanisms in affected regions of brain. We have showed that the concentration of cholesterol, cholesteryl palmitate, phosphatidylinositol, phosphatidylserine, phosphatidylethanolamine, sphingomyelin, ganglioside, glyceryl tripalmitate and sulfatide changes in the PVT region of PTSD compared to control rats. A higher concentration of cholesterol suggests a higher level of corticosterone in the brain. Moreover, concentration changes of phospholipids and sphingolipids suggest the alteration of phospholipase A2 (PLA2) which is associated with inflammatory processes in the brain. Our results have broadened the understanding of biomolecular mechanisms for PTSD in the PVT region of the brain. This is the first report regarding the application of Raman spectroscopy for PTSD studies. This method has a wide spectrum of applications and can be applied to various other brain related disorders or other regions of the brain.
F. Cao, F. Donnarumma, K.K. Murray, Wavelength-Dependent Tip-Enhanced Laser Ablation of Organic Dyes, Journal of Physical Chemistry C, 124 (2020) 1918-1922; doi: 10.1021/acs.jpcc.9b08081
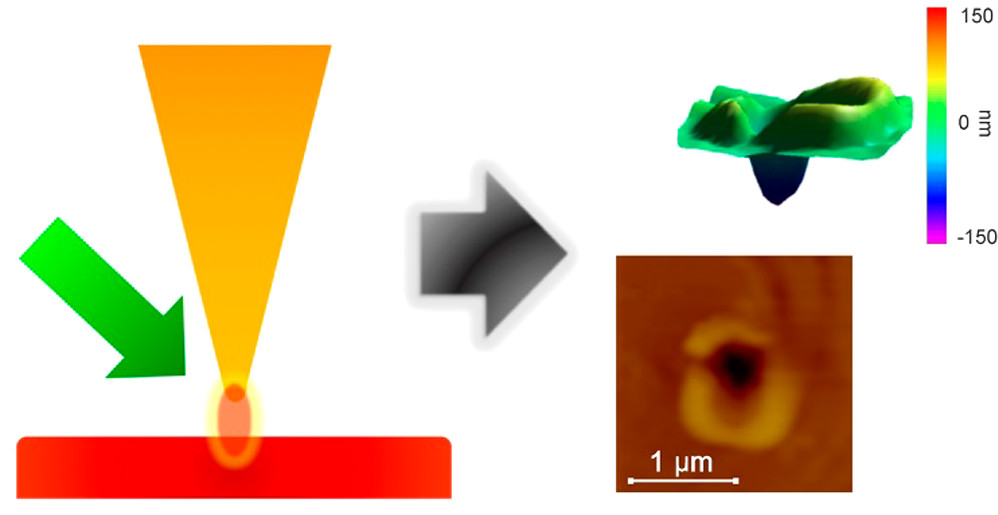
The wavelength dependence of atomic force microscope apertureless tip-enhanced laser ablation was studied using a series of organic dyes to assess the effect of surface optical absorption. An optical parametric oscillator laser with a tunable wavelength range of 410-2100 nm was used to irradiate a gold-coated atomic force microscope tip which was held 15 nm above the surface of a dye thin film vibrating in tapping mode to generate tip-enhanced laser ablation. Crater formation was investigated for rhodamine B, methylene blue, and IR 797 dye thin films which have absorption maxima near 550, 650, and 700 nm, respectively. Crater formation was not observed for wavelengths greater than 700 nm for any of the dyes. Below 700 nm, the crater size was greatest near 500 nm for all dyes and decreased from 500 to 700 nm, and no ablation was observed at longer wavelengths. The crater size did not correlate with the solution or thin film optical absorption of the dyes. The mechanism for tip-enhanced laser ablation is postulated to be either ballistic ejection of gold atoms or direct heat transfer from the tip to the surface.