






Murray Mass Spectrometry Group
Research group of Kermit Murray at Louisiana State University where we use lasers for sampling and imaging and study the chemistry and physics of laser ablation.
Murray Group presentations: https://kermitmurray.com/research/asms-2023/

MP 252 Are Native Conformations of Proteins Preserved Throughout Laser Ablation?; Neda Feizi Gilandeh1; Blessing Chisom Egbejiogu2; Kyle L. Wilhelm1; Kermit K. Murray2; Touradj Solouki1; 1Baylor University, Waco, TX; 2Louisiana State University, Baton Rouge, LA
TP 185 Protein Footprinting Using Water Photolysis; Oluwatosin A. Ogundairo; Kermit K. Murray1; Louisiana State University, Baton Rouge, LA
TP 361 Localized Microsampling of Formalin Fixed Paraffin Embedded Tissue for Proteomics; Blessing Chisom Egbejiogu; Kermit K. Murray; Louisiana State University, Baton Rouge, LA
TP 250 Proteomics of Western Honeybees to Assess Colony Health; Vincent Ricigliono1; Taylar Bell2; Ally Martin1; Fabrizio Donnarumma2; Kermit K. Murray2; 1USDA-ARS, Honey BeecBreeding, Genetics, and Physiology Research, Baton Rouge, Louisiana; 2Louisiana State University, Baton Rouge, LA
WP 428 Deep Ultraviolet Laser Ablation and Capture for Off-line Mass Spectrometry; Kadeem O Hayes1; Blessing Egbejiogu1; Neda Feizi Gilandeh2; Kelcey B. Hines1; Touradj Solouki2; Kermit K. Murray1; 1Louisiana State University, Baton Rouge, LA; 2Baylor University, Waco, TX
WP 456 Deep Ultraviolet Laser Ablation Electrospray Ionization for Native Mass Spectrometry; Kelcey B. Hines1; Neda Gilandeh2; Raul Villacob2; Touradj Solouki2; Kermit K. Murray1; 1Louisiana State University, Baton Rouge, LA; 2Baylor University, Waco, TX
ThP 051 Social Media for Mass Spectrometry; Kermit K. Murray; Louisiana State University, Baton Rouge, LA
MP252
Authors
Neda Feizi Gilandeh1; Blessing Chisom Egbejiogu2; Kyle L. Wilhelm1; Kermit K. Murray2; Touradj Solouki1
Institutes
1Baylor University, Waco, TX; 2Louisiana State University, Baton Rouge, LA
Introduction
Native electrospray ionization mass spectrometry (n-ESI/MS) combined with ion mobility spectrometry (IMS) provides an opportunity to study indigenous forms of proteins in biological samples. Recently, we reported on combining n-ESI/MS with laser ablation sample transfer (LAST) and showed that collision-induced unfolding (CIU) characteristics of proteins before and after LAST were indistinguishable. However, gas-phase CIU results could not conclusively confirm that native structures were preserved throughout the entire laser ablation process. For instance, some “denatured” proteins might fold back to their original native-like forms after solvent capture and complicate the data analysis. Here, we utilize protein denaturing/renaturing, combined with CIU, MALDI, and proteomics results to show proteins retain their original conformations throughout the laser ablation process.
Methods
A custom optic parametric oscillator system (OPO) at Louisiana State University (LSU) was used to laser ablate protein samples and ship them to Baylor University (BU) for native MS and bottom-up proteomics analyses. Stock solution for proteins were adjusted to 200 µM in a mixture of 95% 200 mM NH 4OAc:5% ACN (pH 7.3 ± 0.1). Native IM-MS, UPLC-MS, and broadband collision-induced unfolding data were acquired on a Waters Synapt G2-S system (Milford, MA) at BU. Bottom-up digestion using a single-pot solid-phase-enhanced sample preparation (SP3) method followed by UPLC-IM-MS/MS under enhanced data-independent acquisition (UPLC-HDMS E) was used for protein identification; results were further confirmed by MALDI-MS at LSU. All CIU data were extracted, plotted, and analyzed using CIU Suite 2.
Preliminary Data
As corroborated with our preliminary CIU data, denatured forms of some proteins ( e.g., Bovine Serum Albumin (BSA)) could readily refold back to their native-like structures in ammonium acetate. However, denatured Carbonic Anhydrase (CA) species did not refold back to their original forms in ammonium acetate solvent. We exploited this protein aggregation property of CA along with MALDI, and bottom-up proteomics data to study the preservation of native conformations throughout the laser ablation. Broadband CIU data collected at gas-phase unfolding activation energy intervals of five volts from 10 V to 120 V for nESI and denatured-refolded proteins (in solution) yielded indistinguishable CIU curves. Unlike BSA, denatured CA proteins did not refold to their native structure and seemed to form protein aggregates. For instance, in agreement with previous native-MS reports, CA directly dissolved in native ammonium acetate yielded charge-state species +10, +9, and +8. Once the CA was dissolved in a conventional ESI solvent (~49%:~50%:~0.1% water: ACN: formic acid), denatured CA showed the presence of expected higher charge states (z >+20). However, ESI mass spectrum acquired after drying the denatured CA and redissolving it in a native solvent didn’t show any peaks corresponding to carbonic anhydrase; these results suggest that CA does not refold back to its original native structure. Moreover, the absence of any MS signal for CA in the renaturing solvent is consistent with previous findings and suggests the formation of large protein aggregates that cannot be detected in nESI; our preliminary MALDI and proteomics results are consistent with this view. Our preliminary ion mobility and CIU data for CA before and after the laser ablation yielded indistinguishable plots. Strategies for studying potential conformational changes during the laser ablation process will be presented.
Novel Aspect
Native mass spectrometry combined with bottom-up proteomics confirms preservation of protein conformations throughout the laser ablation process.
ThP 051
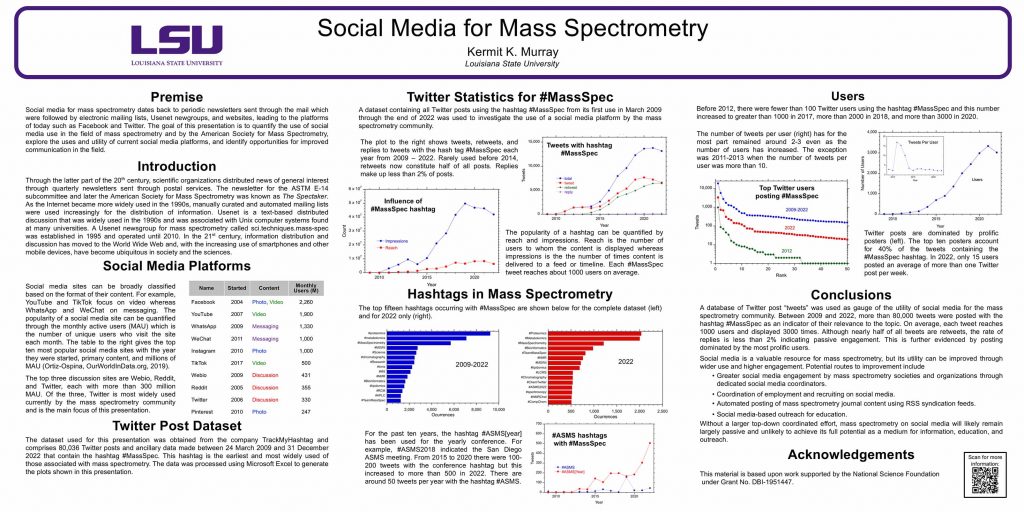
Social media for mass spectrometry dates back to periodic newsletters sent through the mail which were followed by electronic mailing lists, Usenet newgroups, and websites, leading to the platforms of today such as Facebook and Twitter. The goal of this presentation is to quantify the use of social media use in the field of mass spectrometry and by the American Society for Mass Spectrometry, explore the uses and utility of current social media platforms, and identify opportunities for improved communication in the field.
Through the latter part of the 20th century, scientific organizations distributed news of general interest through quarterly newsletters sent through postal services. The newsletter for the ASTM E-14 subcommittee and later the American Society for Mass Spectrometry was known as The Spectaker. As the Internet became more widely used in the 1990s, manually curated and automated mailing lists were used increasingly for the distribution of information. Usenet is a text-based distributed discussion that was widely used in the 1990s and was associated with Unix computer systems found at many universities. A Usenet newsgroup for mass spectrometry called sci.techniques.mass-spec was established in 1995 and operated until 2010. In the 21st century, information distribution and discussion has moved to the World Wide Web and, with the increasing use of smartphones and other mobile devices, have become ubiquitous in society and the sciences.
Social media sites can be broadly classified based on the format of their content. For example, YouTube and TikTok focus on video whereas WhatsApp and WeChat on messaging. The popularity of a social media site can be quantified through the monthly active users (MAU) which is the number of unique users who visit the site each month. The table to the right gives the top ten most popular social media sites with the year they were started, primary content, and millions of MAU (Ortiz-Ospina, OurWorldInData.org, 2019).
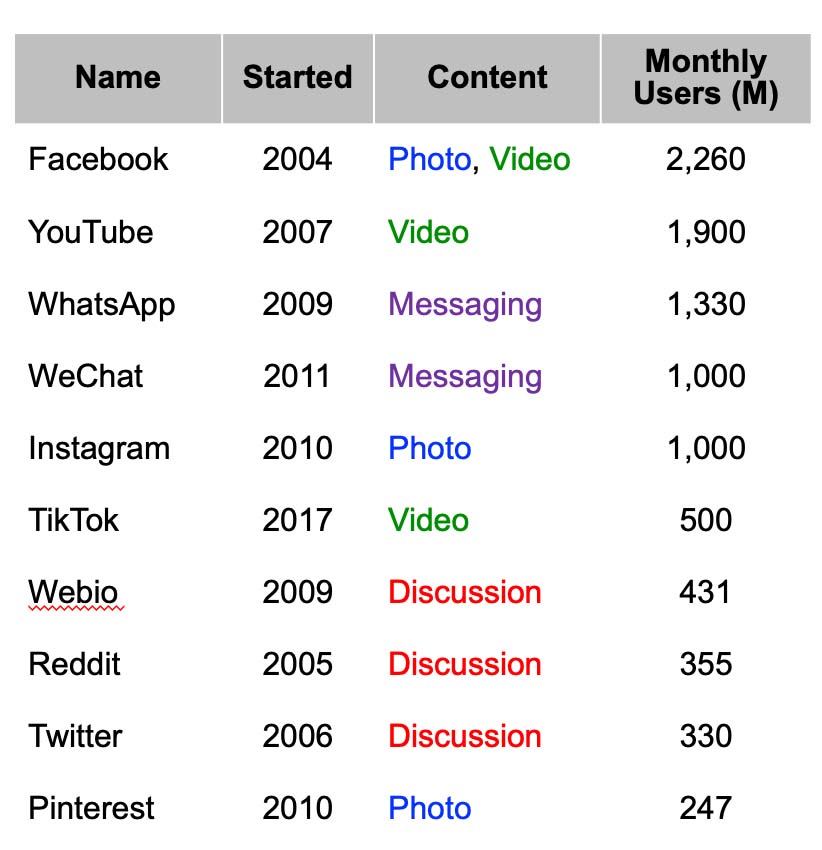
The top three discussion sites are Webio, Reddit, and Twitter, each with more than 300 million MAU. Of the three, Twitter is most widely used currently by the mass spectrometry community and is the main focus of this presentation.
The dataset used for this presentation was obtained from the company TrackMyHashtag and comprises 80,036 Twitter posts and ancillary data made between 24 March 2009 and 31 December 2022 that contain the hashtag #MassSpec. This hashtag is the earliest and most widely used of those associated with mass spectrometry. The data was processed using Microsoft Excel to generate the plots shown in this presentation.
A dataset containing all Twitter posts using the hashtag #MassSpec from its first use in March 2009 through the end of 2022 was used to investigate the use of a social media platform by the mass spectrometry community.
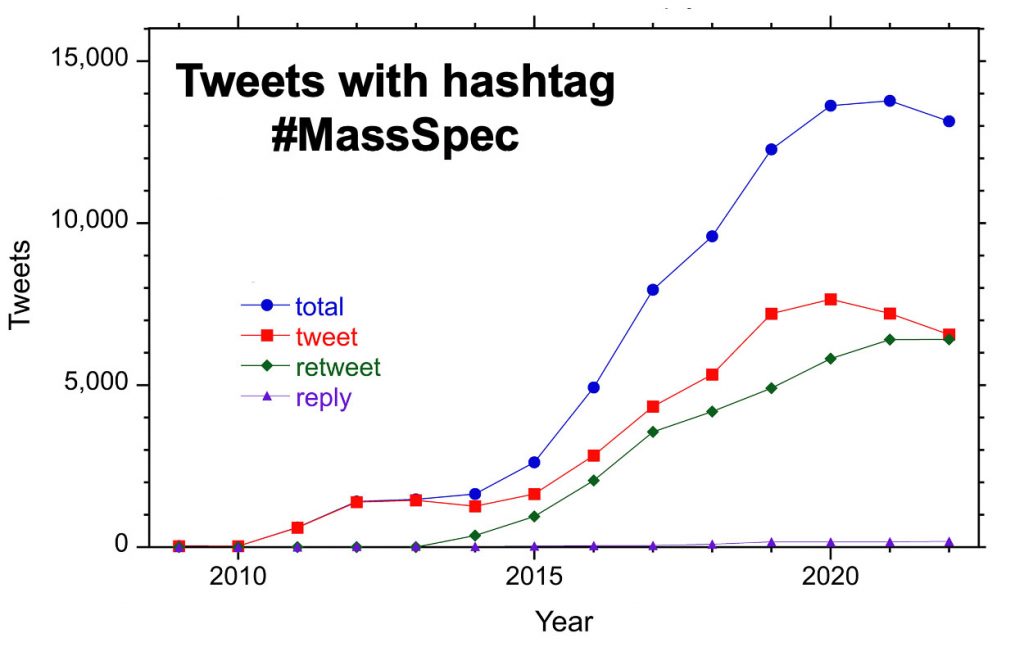
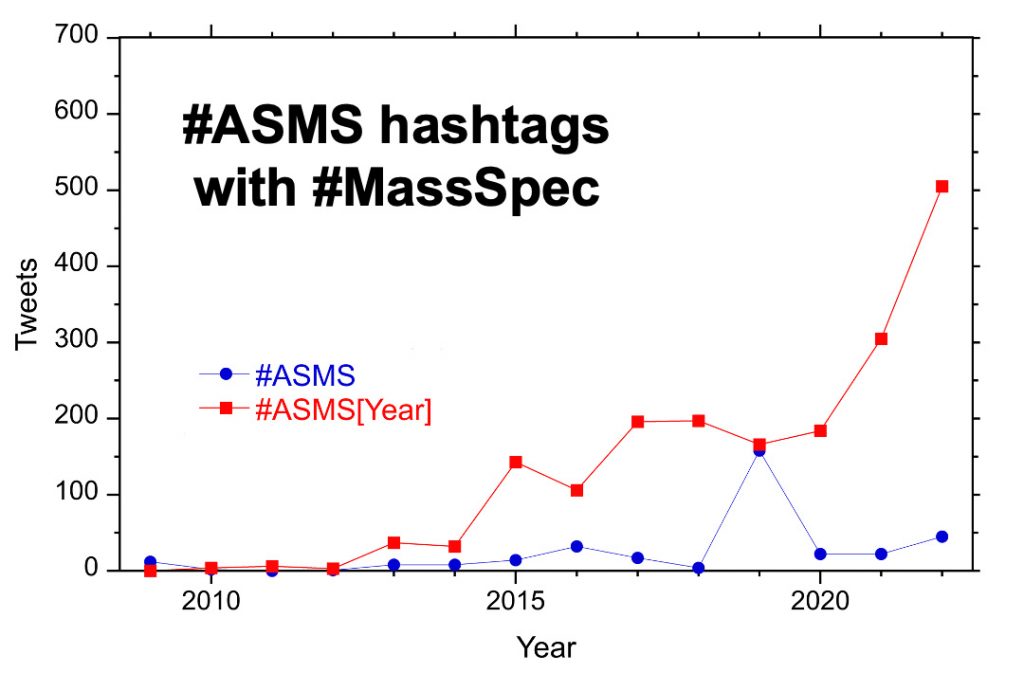
The plot above shows tweets, retweets, and replies to tweets with the hash tag #MassSpec each year from 2009 – 2022. Rarely used before 2014, retweets now constitute half of all posts. Replies make up less than 2% of posts.
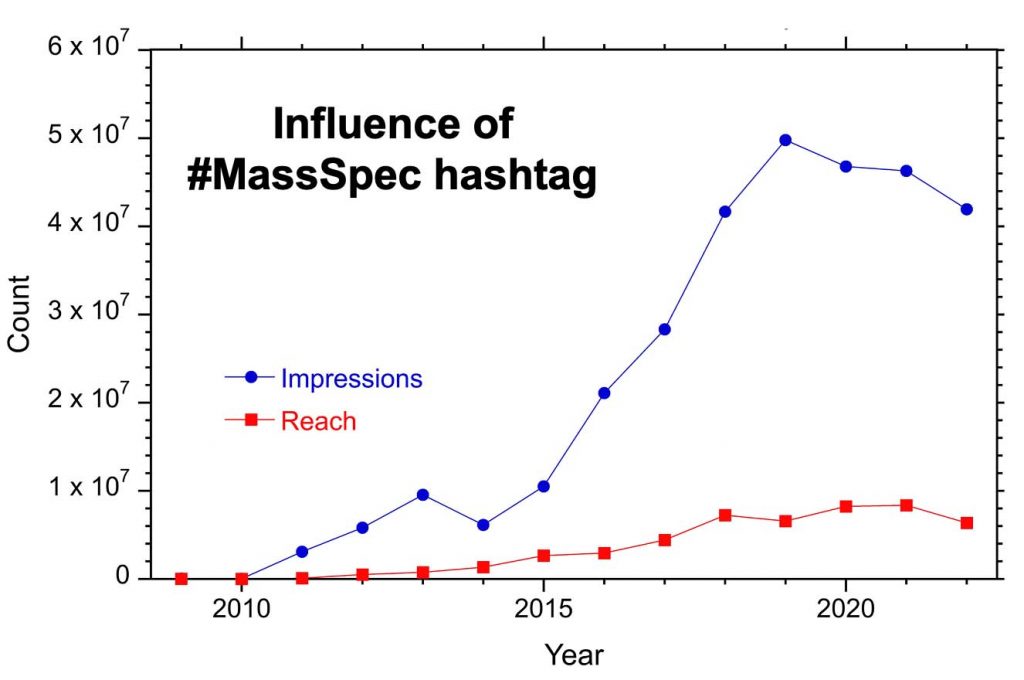
The popularity of a hashtag can be quantified by reach and impressions. Reach is the number of users to whom the content is displayed whereas impressions is the the number of times content is delivered to a feed or timeline. Each #MassSpec tweet reaches about 1000 users on average.
The top fifteen hashtags occurring with #MassSpec are shown below for the complete dataset (left) and for 2022 only (right).
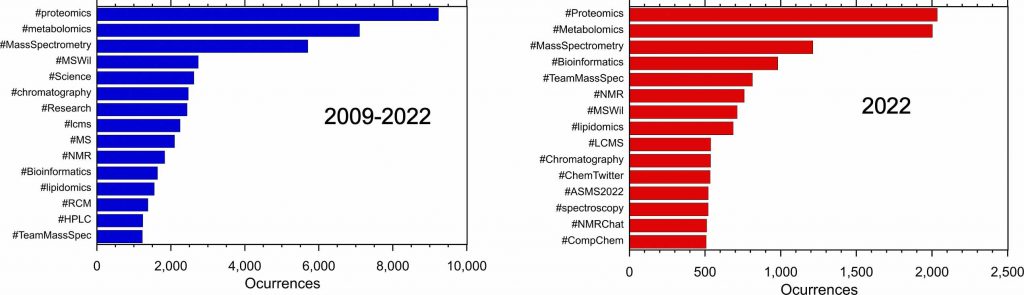
For the past ten years, the hashtag #ASMS[year] has been used for the yearly conference. For example, #ASMS2018 indicated the San Diego ASMS meeting and the 2023 meeting in Houston is represented by #ASMS2023. From 2015 to 2020 there were 100-200 tweets with the conference hashtag but this increased to more than 500 in 2022. There are around 50 tweets per year with the hashtag #ASMS.
Before 2012, there were fewer than 100 Twitter users using the hashtag #MassSpec and this number increased to greater than 1000 in 2017, more than 2000 in 2018, and more than 3000 in 2020.
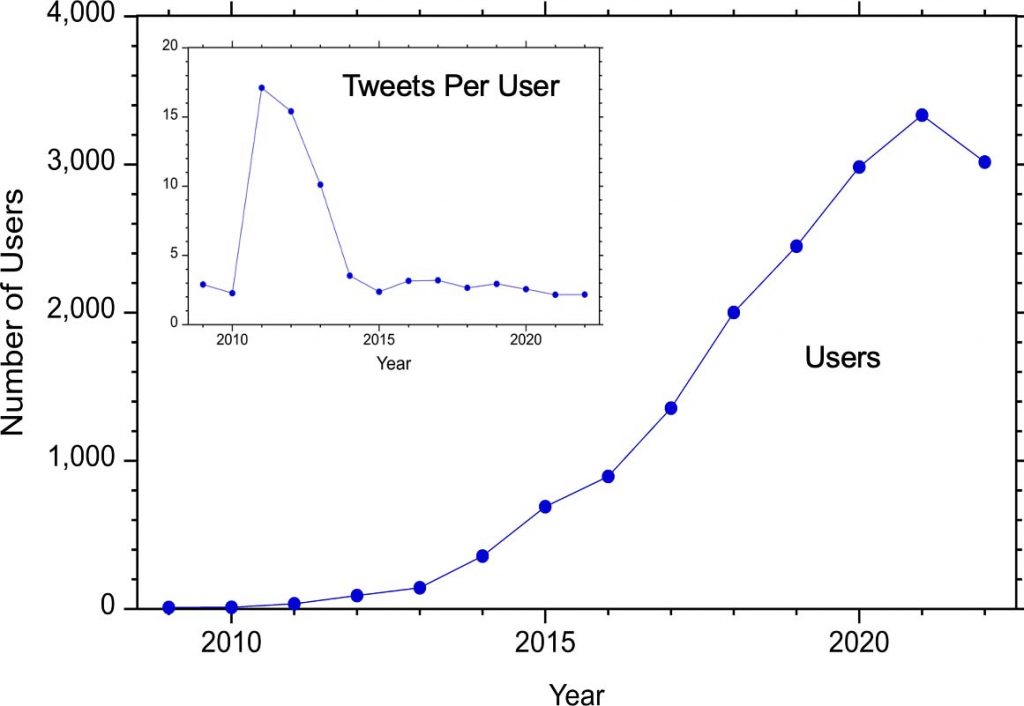
The number of tweets per user (above inset) has for the most part remained around 2-3 even as the number of users has increased. The exception was 2011-2013 when the number of tweets per user was more than 10.
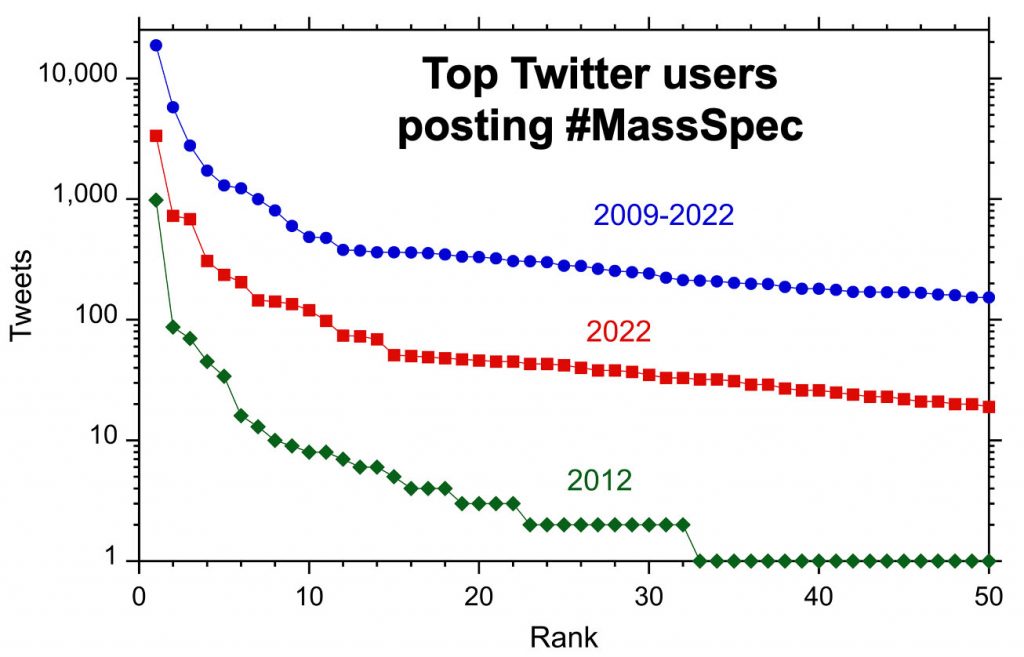
Twitter posts are dominated by prolific posters (above). The top ten posters account for 40% of the tweets containing the #MassSpec hashtag. In 2022, only 15 users posted an average of more than one Twitter post per week.
A database of Twitter post “tweets” was used as gauge of the utility of social media for the mass spectrometry community. Between 2009 and 2022, more than 80,000 tweets were posted with the hashtag #MassSpec as an indicator of their relevance to the topic. On average, each tweet reaches 1000 users and displayed 3000 times. Although nearly half of all tweets are retweets, the rate of replies is less than 2% indicating passive engagement. This is further evidenced by posting dominated by the most prolific users.
Social media is a valuable resource for mass spectrometry, but its utility can be improved through wider use and higher engagement. Potential routes to improvement include
Without a larger top-down coordinated effort, mass spectrometry on social media will likely remain largely passive and unlikely to achieve its full potential as a medium for information, education, and outreach.
This material is based upon work supported by the National Science Foundation under Grant No. DBI-1951447.
I set up an account on a Mastodon instance: mastodon.online/@kkmurray
Good hash tags seem to be #massspec and #teammassspec