A.C. Pulukkody, Y.P. Yung, F. Donnarumma, K.K. Murray, R.P. Carlson, L. Hanley, Spatially resolved analysis of Pseudomonas aeruginosa biofilm proteomes measured by laser ablation sample transfer, PLoS One, 16 (2021) e0250911, doi: 10.1371/journal.pone.0250911
Abstract
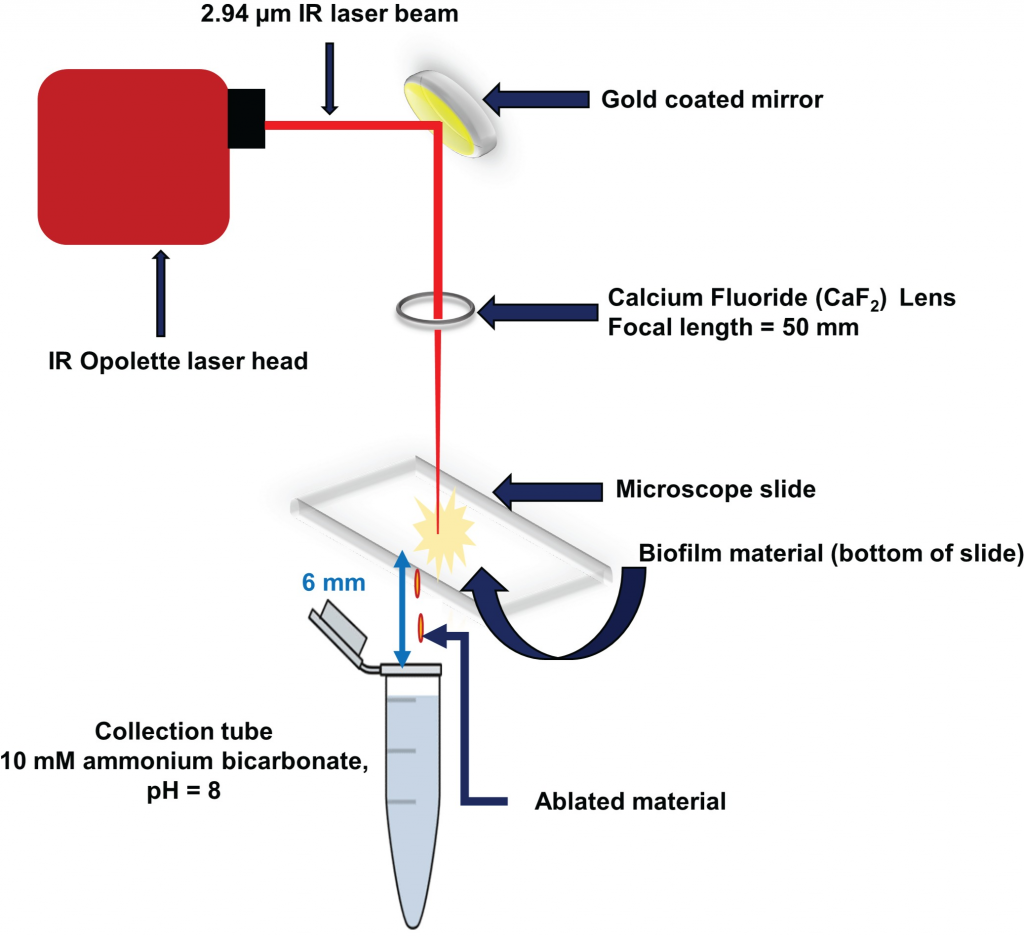
Heterogeneity in the distribution of nutrients and oxygen gradients during biofilm growth
gives rise to changes in phenotype. There has been long term interest in identifying spatial
differences during biofilm development including clues that identify chemical heterogeneity.
Laser ablation sample transfer (LAST) allows site-specific sampling combined with label
free proteomics to distinguish radially and axially resolved proteomes for Pseudomonas aeruginosa
biofilms. Specifically, differential protein abundances on oxic vs. anoxic regions of a
biofilm were observed by combining LAST with bottom up proteomics. This study reveals a
more active metabolism in the anoxic region of the biofilm with respect to the oxic region for
this clinical strain of P. aeruginosa, despite this organism being considered an aerobe by
nature. Protein abundance data related to cellular acclimations to chemical gradients
include identification of glucose catabolizing proteins, high abundance of proteins from arginine
and polyamine metabolism, and proteins that could also support virulence and environmental
stress mediation in the anoxic region. Finally, the LAST methodology requires only a
few mm2 of biofilm area to identify hundreds of proteins.

